Initial Genome-Wide Association
Study of Feed Intake-Related
Traits in Beef Cattle
Feed Efficiency Consortium
http://www.beefefficiency.org/info/FE_Genomics_FactSheet.pdf
*Click here for printable pdf version.
Importance of Feed Efficiency
The ability to improve the utilization of nutrients has tremendous potential to improve profitability. It has been estimated that a 10% improvement in performance (gain) would increase profit by 18%, while a 10% improvement in efficiency could improve profit by upwards of 43%. It has also been estimated that a 10% improvement in feed efficiency (assumed to be a 2 lb/day reduction in RFI) across the entire feedlot sector would equate to $1.2 billion in reduced feed costs.
Although progress has been made in feed conversion (F:G) over the past decade, it has been small relative to the progress of other species. The poultry industry, for instance, has made a 250% increase in feed efficiency since 1957. Iowa closeout data suggest a 0.047 lb/yr decrease in F:G from 1978-1992 and from 1988-2002 Midwestern closeout data suggest that the change has been slightly less (0.033 lbs/yr decrease). Advancements in dietary regimes and technology (implants and feed additives) have made substantial differences, but direct genetic selection for efficiency remains an untapped source of potential improvement.
Why a Genomics Approach?
Genomic-enhanced EPD were first estimated for carcass traits and then evolved to other production traits for which EPDs already existed. These traits were chosen due to the need for phenotypes to develop the genomic predictions (training), which were already being collected. Consequently, genomic tests for “novel” traits such as different measures of efficiency or disease susceptibility require a significant effort in order to build large resource populations of animals with both phenotypes and genotypes. These two particular suites of traits (Feed Efficiency and Bovine Respiratory Disease) are currently the foci of two integrated USDA projects. In these two cases, use of genomic tools could have an economic advantage over routine collection of costly phenotypes.
Current Efforts and Results
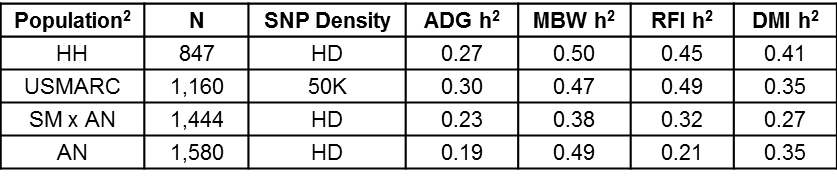
A multi-institutional integrated research, extension and educational effort to develop and deploy selection tools to improve the efficiency of feed utilization in growing cattle is currently underway (www.beefefficiency.org). Since feed intake phenotypes are expensive to measure and a genomics approach is logical, this project seeks to develop genomic predictors for feed intake/efficiency using dense single nucleotide polymorphism (SNP) panels (50,000 and 770,000 SNPs). The project also plans to dissect regions of the bovine genome that harbor genetic variants that explain relatively large portions of the genetic variation for these traits in an effort to discover the genes that control the underlying mechanisms that make animals more efficient. To do this requires the collection of feed intake records from thousands of animals that are genotyped with either the 50K or 770K (HD) SNP assays across multiple breeds in order to develop genomic predictors that are accurate and robust across cattle populations. Table 1 contains initial results from the USDA project National Program for Genetic Improvement of Feed Efficiency in Beef Cattle. Four populations of cattle involving over 5,000 animals were used for a genome-wide association study (GWAS) of different feed efficiency related traits. The heritability estimates (h2) represent the proportion of phenotypic variation explained by the SNPs. Although all estimates suggest that these traits are moderately heritable, differences in parameter estimates exist between the resource populations likely due to differences in population size, structure (e.g., the number of contemporary groups), and data collection methods. Some of the Angus (AN) animals for instance were fed in Calan gates whereas the other populations were fed via automated feed intake systems (e.g., GrowSafe).
|
Table 1. Genomic heritability estimates for ADG, MBW, RFI and DMI1 |
 |
|
1 ADG = Average daily gain, MBW = mid-test metabolic body weight, RFI = Residual feed intake, DMI = dry matter intake. 2 HH=Hereford cattle fed at Olsen Ranches, USMARC=F12 composites from the Meat Animal Research Center Cycle VII, SM x AN=Legacy Simmental x Angus animals fed at the University of Illinois, AN=Angus cattle fed at Circle A and the University of Missouri. |
Results from the GWAS also identified regions of the genome that appear to harbor large effect quantitative trait loci (QTL). Given the complex nature of these traits (the fact they are controlled by numerous genes), a large effect QTL was considered as a locus explaining greater than 1% of the additive genetic variation. A total of 5, 5, 17, and 10 of these large effect QTL were identified for ADG, DMI, MBW and RFI, respectively. Some of the QTL identified had substantially larger effects than might have been expected. For instance, a QTL in Angus explained over 10 and 14% of the additive genetic variation in DMI and MBW, respectively. No QTL identified for RFI explained greater than 2.5% of the additive genetic variation. These QTL regions were generally breed specific, further illustrating why genomic predictors are not easily transferable across breeds.
Summary
Results from the current study illustrate that by using either the 50K or 770K SNP assay, the genomic heritability estimates of traits related to feed efficiency are in general agreement with heritability estimates from the scientific literature using phenotypes and pedigree information. The fact that these traits are moderately heritable and that the SNP assays can explain large proportions of the phenotypic variation suggest that genetic progress in these traits can be made by using genomic selection. However, this study further illustrates the breed specific nature of genomic predictors and thus caution should be used if attempting to use a genomic predictor in a population that is distantly related to the training population (e.g., across breeds). The continued collection of feed intake phenotypes will be required to refine and retrain genomic predictions overtime.
The ability to select for improved feed utilization is exciting and will be enabled by genomic tools. However, the improvement of efficiency is inherently a multiple-trait issue and thus the development and utilization of indexes to select for the most profitable animals is critical. An interactive example of such an index is available at www.beefefficiency.org.
Full research article reference:
Mahdi Saatchi, Jonathan E Beever, Jared E. Decker, Dan B. Faulkner, Harvey C Freetly, Stephanie L Hansen, Helen Yampara-Iquise, Kristen A Johnson, Stephen D Kachman, Monty S Kerley, JaeWoo Kim, Daniel D Loy, Elisa Marques, Holly L Neibergs, E John Pollak, Robert D Schnabel, Christopher M Seabury, Daniel W Shike, Warren M Snelling, Matthew L Spangler, Robert L Weaber, Dorian J Garrick and Jeremy F Taylor. 2014. QTL, candidate genes, metabolic and signaling pathways associated with growth, metabolic mid-test weight, feed intake and feed efficiency in beef cattle. BMC Genomics.

