
How DNA Testing Will Affect the
Accuracy of EPD Information
Bob Weaber, Kansas State University
bweaber@k-state.edu
Matthew Spangler, University of Nebraska
mspangler2@unl.edu
*Click here for printable pdf version.
Summary: Genomic information can increase EPD accuracy, particularly for non-parent animals. Collecting phenotypes is still critical.
Introduction
Selection decisions in the beef industry have been fostered by the development and delivery of Expected Progeny Differences (EPD) for a wide variety of traits and across all major US beef breeds. Since the early 1970’s, EPDs have been used by seedstock and commercial beef producers to make genetic change in their herds. Today, EPDs are widely accepted across the industry and are used frequently by producers making seedstock selection and purchase decisions. The degree of confidence in an individual animal’s EPDs is described numerically by a computed value called ‘Accuracy.’ Accuracy values in the US are scaled reliabilities and range from 0 to 1 representing the amount of information used to compute the EPD. An animal with accuracy values near zero has very little data available for evaluation while an animal with accuracy of 0.99 has a very large amount of information evaluated.
Improvements in EPD accuracy have historically been driven by phenotypic record collection directly on the trait of interest or on indicator traits. For traits like stayability or length of productive life, the evaluation of a sire’s daughters is typically completed long after the bull has been removed from production. For other traits like carcass weight, marbling score, and rib-eye area, the animal must be harvested or ultrasound information collected as indicator trait data. There are costs associated with collecting and processing phenotypic data. To achieve high levels of accuracy a great deal of progeny and/or grand progeny data must be included in the evaluation.
Timing is Everything
Accuracy values for bulls purchased by commercial producers as yearlings will be low. In most cases only the bull’s own performance records for traits observed before sale day and pedigree information will be included in his EPD calculations. For the maternal traits like heifer pregnancy, stayability and maternal milk no daughters will have been produced so only pedigree estimate or interim EPDs will be available, and these EPD have low accuracies. In order to improve the accuracy of the EPDs of yearling bulls another source of information is needed.
Genomic information gives an accurate picture of what alleles an offspring inherited from its parents in the form of Single Nucleotide Polymorphisms (SNP), and has always held the promise to increase the accuracy of EPD. This promise has finally been realized for those breeds that had breed-specific training populations that enable genotypic information to be translated into genetic merit estimates (i.e. Molecular Breeding Values (MBV)) that can be incorporated into genomic-enhanced EPD calculations. Studies have shown that genomic information cannot be accurately translated into MBV for complex traits (i.e. those controlled by many genes) in the absence of breed-specific training populations.
One key advantage of MBV is that this information can be garnered early in the life of the animal thus enabling an increase in the accuracy of EPD particularly on young animals, which have not yet produced progeny. Ideally, MBV data should be used to increase the accuracy of the EPDs of young animals prior to any selection decisions (performance based culling) made at the seedstock level. Seedstock genetic trends and subsequent genetic flow to commercial producers will only be improved if seedstock producers actually use the genomic-enhanced EPDs to make selection decisions for animals that will be retained as breeding animals and offered for sale to commercial producers. Genotyping a group of animals immediately before sale after all selection has been completed does nothing to improve genetics of the population; it only fosters marketing efforts and only allows for better selection decisions within a highly selected subset of the sale offering.
The US Beef Industry has witnessed considerable evolution in terms of the genomic tests available in the market place. The tests that are currently being included in EPD are comprised of 50,000 (50K) SNP, although some breeds utilize 80K panels and some are moving towards reduced (e.g. 20K) panels with the aid of imputation (essentially using information from the population to “replace” missing genotypes). The research community is commonly using 50K, 80K or 770K genomic tests for discovery of “novel” traits (i.e. feed efficiency, disease susceptibility).
Implementation
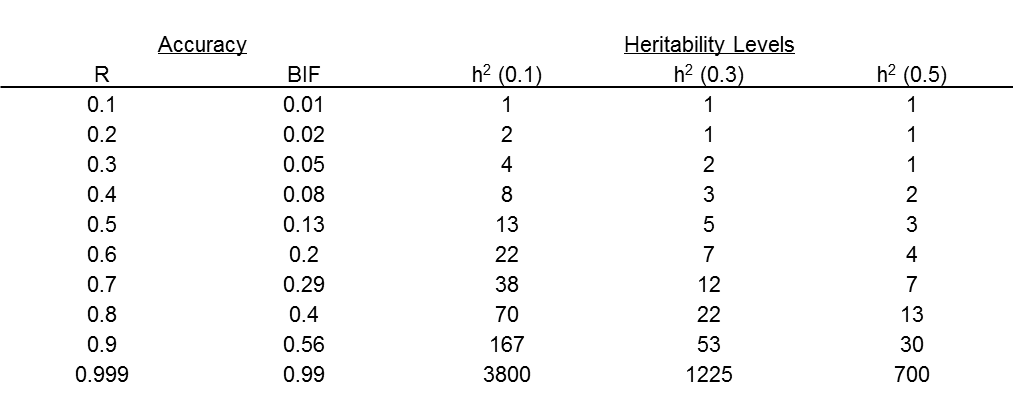
The underlying question commonly asked by producers is “Do genomic tests work?” It is critical to understand that this is a somewhat ambiguous question, as the true answer is not binary (i.e. yes or no). The important question to ask is “How well do genomic tests work?”, and the answer to that question is related to how much of the genetic variation the genomic test explains. The benefit will be dependent upon the proportion of genetic variation (%GV) explained by a given genomic test. The %GV is equal to the square of the genetic correlation multiplied by 100. Table 1 shows the relationship between the genetic correlations (true accuracy), %GV and Beef Improvement Federation (BIF) accuracy. BIF accuracy is the standard for all U.S. beef breeds.
|
Table 1. The relationship between true accuracy (r), proportion of genetic variation explained (%GV), and Beef Improvement Federation (BIF) accuracy. |
 |
Molecular Breeding Values should not be thought of as a separate independent predictor of genetic merit, but rather as a potentially useful indicator that is correlated to the trait of interest. Combining the genomic information with traditional sources of EPD information increases the accuracy of the resulting genomic-enhanced EPD and this has the potential to increase the rate of genetic change by both increasing the accuracy of selection, and decreasing the generation interval. This latter component of the breeder’s equation would be particularly impacted if young sires are used more frequently as a result of the increased confidence in their genetic superiority due to added genomic information.
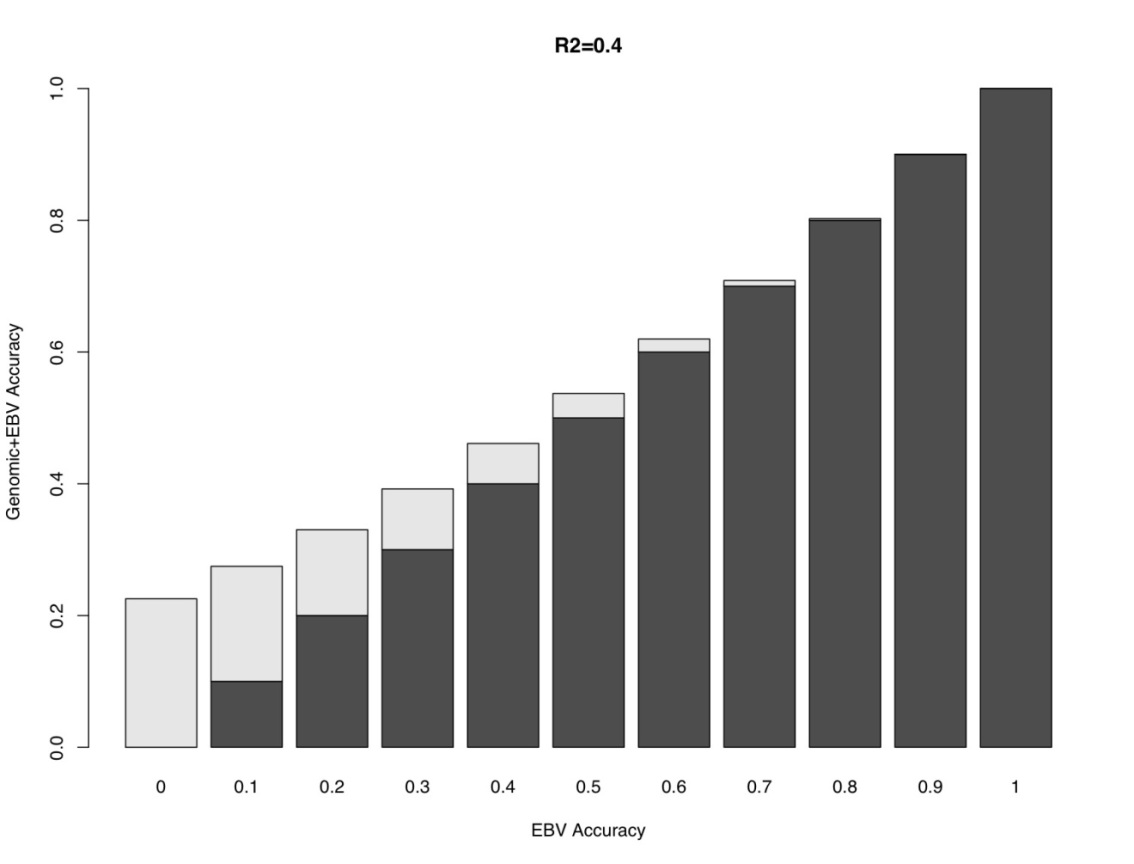
Figure 1 illustrates the benefit of incorporating genomic information into a genomic-enhanced EPD on accuracy (on the BIF scale) when the MBV explains 40% of the genetic variation (GV), which is synonymous with an r2 value of 0.4. The darker portion of the bars shows the EPD accuracy before the inclusion of genomic information and the lighter colored portion shows the increase in accuracy after the inclusion of the MBV into the EPD calculation. As the %GV increases, the increase in EPD accuracy becomes larger. Additionally, lower accuracy animals benefit more from the inclusion of genomic information and the benefits decline as the EPD accuracy increases. Regardless of the %GV assumed here, the benefits of including genomic information into EPD dissipate when EPD accuracy is between 0.6 and 0.7. On the other hand, when %GV is 40, an animal with 0 (zero) accuracy could exceed 0.2 accuracy with genomic information alone. This would be comparable to having approximately 4 progeny for a highly heritable trait or 7 progeny for a moderately heritable trait (Table 2).
 |
|
Figure 1. Increase in accuracy from integrating genomic information that explains 40% of the genetic variation into Estimated Breeding Values (EBV). |
|
Table 2. Approximate number of progeny needed to reach accuracy levels (true (r) and the BIF standard) for three heritabilities (h2). |
 |
Although the American Simmental Association (ASA) was the first to augment their Warner Bratzler Shear Force EPD with genomic information, several other breeds have adopted this technology and others are in the process of collecting sufficient records to develop breed-specific training populations. Research has shown moderate to high genetic correlations between several traits of interest and MBV in multiple breeds when the animals the test is used on are within the same breed as the training data set used to develop the MBV. However, it has also been clearly demonstrated that when a MBV developed in one breed is used in a different breed, even a closely related breed (e.g. Angus and Red Angus), the genetic correlation drops substantially.
This shows the unfortunate breed specificity issues surrounding these tools. This is consistent with other results that show the predictive power of MBV begin to erode as the genetic distance between the training and target (or evaluation) populations increase. This would be expected overtime as animals in the training data used to develop the MBV become more distantly related to animals currently being evaluated with the genomic test. This is why these tools need to be “re-trained” or “re-calibrated” periodically.
Some breeds do not have the luxury of immediately having thousands of genotyped animals for use in developing breed-specific training populations. Consequently, the use of a robust across-breed set of genomic prediction equations would be beneficial. There are two primary methods of constructing an across-breed training data set: pool purebred animals from multiple breeds or use crossbred animals. The first option requires the use of EPD, corrected for differences in accuracy, as “phenotypes” for training similar to the within breed scenario with the exception of correcting for breed effects in the model. The second option requires the use of adjusted phenotypes (corrected for contemporary group effects, sex, etc.) to train the genomic predictors. Although pooling animals of different breed together in training can be useful, it only helps if it will be used in breeds that were represented in the training data.
Conclusions
Genomics and the corresponding Marker-Assisted or Genomic-Enhanced EPD, have become a reality. Within-breed genomic predictions based on 50K genotypes have proven to add accuracy, particularly to young bulls, for several traits. The push going forward will be the adoption of this technology by other breed associations. Furthermore, methodology related to the use of this technology in crossbred or composite cattle is critically needed, . The crux of adoption will be getting commercial bull buyers to see the value in, and thus pay for, increased EPD accuracy. There is still a need to collect and routinely record phenotypic information by seedstock producers. Commercial producers need to realize that EPDs, and economic index values, are the currency of the realm for beef cattle selection. Genomic technology only makes these tools stronger, it does not replace them.
